M3: A new platform for elucidation of large BIOMOLECULAR complexes
Life is operated at the nanometer scale through orchestrated communications of biomolecules. By solving the structures of this nanoworld, we can acquire a fundamental understanding of how biological macromolecules function, how they are related to disease-linked pathways and how to design drugs targeting them.
Together with her colleagues at EMBL, Dr. Ezgi Karaca who joined IBG recently, has developed an integrated structure determination framework M3 (Model Molecular Machines) that is able to simply and effectively calculate the structure of large protein-RNA complexes on the basis of diverse experimental data. “The data can, for example, come from mutational analyses, NMR spectroscopy, electron microscopy, mass spectrometry, or evolutionary conservation,” explains Prof. Teresa Carlomagno (Helmholtz Centre for Infection Research), the team leader of the M3 project (Figure 1). “By combining all methods, a calculation of the protein complex is possible, which gives us a first idea about what we are faced with. At the same time, the M3 platform also provides scientists with information when data are not yet sufficient and further experiments are necessary to accurately describe the protein complex under study.” These important news have been shared with the scientific community by Helmholtz Centre for Infection Research (https://www.helmholtz-hzi.de/en/news_events/news/view/article/complete/easier_decoding_of_unknown_protein_complexes/) and University of Utrecht (https://www.uu.nl/en/news/new-platform-for-elucidation-of-large-protein-and-nucleic-acid-structures-in-infections), respectively.
The M3 tool is available free of charge for non-profit researchers. The researchers published their results in the renowned scientific journal Nature Methods.
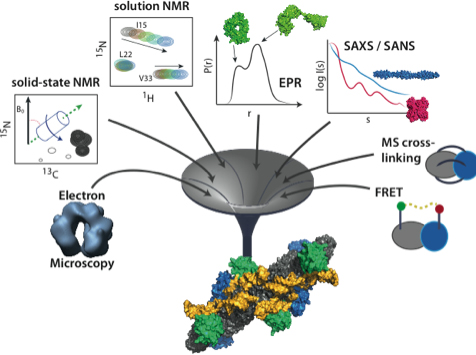 |
| Figure 1: M3 can combine experimental data from diverse sources, in order to determine the structures of molecular machines. |
M3 FRAMEWORK
The M3 Framework is a broadly applicable and user-friendly platform, which effectively integrates single and very mixed structural and biochemical data of different origins and calculates the atomic structure of large complex molecular structures. The protocol is based on the HADDOCK software developed by Prof. Alexandre Bonvin (Utrecht University). The additional files required to run the M3 Framework in combination with HADDOCK are publicly available for download: https://github.com/ezgikaraca/ISD-files.
PUBLICATION
Ezgi Karaca, João P.G.L.M. Rodrigues, Andrea Graziadei, Alexandre M.J.J. Bonvin, Teresa Carlomagno. M3: an integrative framework for structure determination of molecular machines. Nature Methods, 2017, DOI: 10.1038/nmeth.4392.

